Barry Sly-Delgado presented our overview paper on TaskVine at the Workshop on Workflows in Support of Large Scale Science at Supercomputing 2023 in Denver, Colorado.
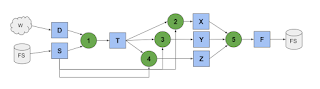
TaskVine is a system for executing data intensive workflows on large clusters. These workflows ma consist of thousands to millions of individual tasks that are connected together in a graph structure like this:
When executing this sort of workflow in a cluster, the movement of data between tasks is often the primary bottleneck, especially if each item must flow back and forth between a shared filesystem.
The key idea of TaskVine is to exploit the local storage available on each node of a cluster to relieve much of the load placed on a shared filesystem. A running TaskVine system consists of a manager process that coordinates data transfers between workers, like this:
A workflow in TaskVine is expressed in in Python by declaring the data assets needed by the workflow as file objects, and then connecting them to the namespace of each task to execute:
When the tasks execute on each worker, the various data objects are cached on local storage, and then linked into the execution sandbox of each running task. This provides the opportunity for data to be shared between tasks without moving data to/from the shared filesystem:
This paper gives three examples of applications constructed using TaskVine. TopEFT is a physics data analysis application built using Coffea+TaskVine, Colmena-XTB is a molecular modeling application built using Parsl+TaskVine, and BGD is a model training application built using TaskVine alone. Each of these codes is able to scale to 400-1000 nodes running on 1000-27000 cores total.
For all the details, please check out our paper here:
- Barry Sly-Delgado, Thanh Son Phung, Colin Thomas, David Simonetti, Andrew Hennessee, Ben Tovar, Douglas Thain, TaskVine: Managing In-Cluster Storage for High-Throughput Data Intensive Workflows, 18th Workshop on Workflows in Support of Large-Scale Science, November, 2023. DOI: 10.1145/3624062.3624277




No comments:
Post a Comment